Abstract
Introduction
AML suppresses immune responses directly by expressing cytokines and immune checkpoint ligands as well as indirectly, by recruiting suppressive immune cells like regulatory T cells (Tregs). Prior studies have looked at a small number of immune checkpoint receptors and ligands on limited subsets of T cells and AML blasts, respectively. The frequency of circulating Tregs has been associated with induction failure in AML (Szczepanski et al CCR 2009) and the coexpression of PD1 and TIM3 on CD8 T cells has been associated with immune exhaustion (Zhou et al Blood 2011) and relapse following allogeneic stem cell transplant (Kong et al Blood Can J).
Recent clinical trials have demonstrated encouraging response rates and durable responses in patients with relapsed AML treated with PD-1 based therapies (Daver et al EHA 2017) and following allogeneic stem cell transplantation in relapsed AML with CTLA-4 based therapies (Davids et al NEJM 2016). Better defining the immune checkpoint landscape of human AML will facilitate the rational selection of immune checkpoint targets and combination approaches for future clinical trials.
Methods
Multi-parameter flow-cytometry assay were performed by Immunotherapy Platform on bone marrow (BM) aspirates from 107 patients with AML treated at the MD Anderson Cancer Center. We evaluated the expression of inhibitory (PD1, CTLA4, LAG3, TIM3) and stimulatory receptors (GITR, OX40, 41BB, ICOS) on T cell subsets: CD4 T effector cells [Teff]: CD3+CD4+CD127lo/+Foxp3-, CD4 T regulatory cells [Treg]: CD3+CD4+CD127-Foxp3+, and CD8 T cells and the expression of their ligands (41BBL, B7-1, B7-2, ICOSL, PDL1, PDL2 and OX40L) on AML blasts. Eight healthy donor BMs were used as controls. We correlated the expression of these markers on T cell subsets and blasts, with age, karyotype and a baseline next generation sequencing for 28 myeloid-associated genes including P53, DNA methylation proteins (DNMT3a, IDH1, IDH2, TET2) and FLT3.
Results
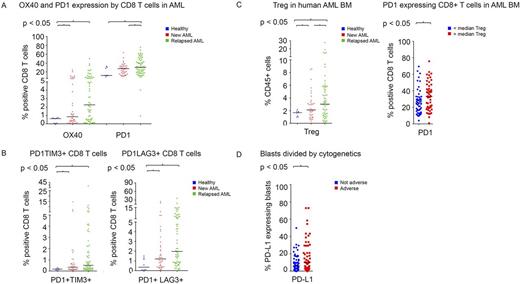
BM aspirates from AML patients demonstrated significantly higher frequency of CD45 gated CD3+ total T cell infiltrate compared to healthy donors. The proportion of CD8+ T cells expressing PD1 was higher in patients with relapsed (32.8%) and with newly diagnosed AML (26.4%) compared to healthy donors (18.5%) (P<0.01 and P=0.03, respectively)(Figure 1A). The proportion of CD8+ T-cells expressing OX40 was also higher in patients with AML as compared to healthy donors. A similar pattern was noted for PD1 and OX40 was noted on the other CD4+ T cell subsets.
Compared with healthy donors, patients with relapsed and newly diagnosed AML had significantly higher percentage of CD8+ T cells co-expressing the inhibitory markers PD1 with TIM3 (P=0.02) and PD1 with LAG3 (P<0.01) (Figure 1B). The frequency of Tregs in BM increased progressively from healthy donors to newly diagnosed AML to relapsed AML (1.6% vs 2.8%, P<0.01, vs 4.5% P<0.01). An increased Treg infiltration correlated with higher proportion of CD8+ T cells expressing PD1 (33.6% vs 27%; P=0.03) (Figure 1C). Patients with increased Treg and PD1+CD8+ T cell infiltration also showed significantly higher PD-L1/L2 expressing AML blasts in their BM.
We attempted to identify potential relationships between molecular characteristics and immune checkpoint expression and found that BM blasts in patients with adverse karyotype more frequently expressed PD-L1 as compared to patients with non-adverse karyotype (17.5% vs 8.8%, P=0.03) (Figure 1D).
Conclusions
Increased Treg and PD1+ CD8 T cell infiltration in AML and the increased PD1+TIM3+ and PD1+LAG3+ CD8 T cells in the BMs of patients with AML, point to worsening exhaustion of an adaptive immune response. The correlation between increased Treg infiltration, increased PD1+CD8 cells and increased PD-L1 expression on blasts in the BM indicates immune suppression at multiple levels. The association between adverse cytogenetics and higher PD-L1 expression on BM blasts further suggests links between AML biology and immune suppression. The dual contributions by T cells and blasts to immune suppression indicate that these pathways may play an important role in AML survival and therefore patients may benefit from checkpoint antibody therapy.
Cortes: ImmunoGen: Consultancy, Research Funding; Novartis Pharmaceuticals Corporation: Consultancy, Research Funding; BMS: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; ARIAD: Consultancy, Research Funding; Sun Pharma: Research Funding; Teva: Research Funding. Allison: Merck: Patents & Royalties: Owns patent licensed to Merck; BMS: Consultancy, Patents & Royalties; Jounce: Consultancy, Patents & Royalties: Owns patent licensed to Jounce; Jounce: Consultancy, Other: Owns stock; Kite Pharma: Consultancy, Other: stock; Evelo: Consultancy, Other: stock; Neon: Consultancy, Other: stock; Constellation: Consultancy, Other: stock; GSK: Consultancy; Astra Zeneca: Consultancy; Amgen: Consultancy; EMD Serono: Consultancy; Astellas: Consultancy. Kantarjian: Delta-Fly Pharma: Research Funding; ARIAD: Research Funding; Pfizer: Research Funding; Amgen: Research Funding; Bristol-Meyers Squibb: Research Funding; Novartis: Research Funding. Sharma: Jounce: Consultancy, Other: stock, Patents & Royalties: Patent licensed to Jounce; Astellas: Consultancy; EMD Serono: Consultancy; Amgen: Consultancy; Astra Zeneca: Consultancy; GSK: Consultancy; Consetellation: Other: stock; Evelo: Consultancy, Other: stock; Neon: Consultancy, Other: stock; Kite Pharma: Consultancy, Other: stock; BMS: Consultancy. Daver: Karyopharm: Consultancy, Research Funding; Novartis Pharmaceuticals Corporation: Consultancy; Immunogen: Research Funding; Incyte Corporation: Honoraria, Research Funding; Pfizer Inc.: Consultancy, Research Funding; Otsuka America Pharmaceutical, Inc.: Consultancy; Jazz: Consultancy; Sunesis Pharmaceuticals, Inc.: Consultancy, Research Funding; Kiromic: Research Funding; Daiichi-Sankyo: Research Funding; Bristol-Myers Squibb Company: Consultancy, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal